ECoG quality checks¶
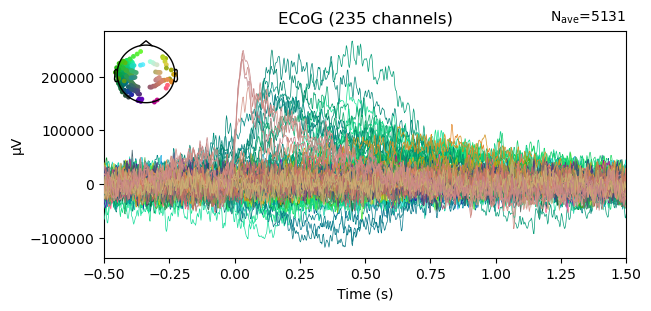
This tutorial will showcase two ways to quality check the dataset. First, we will correlate the podcast audio with each electrode’s time series. We expect that electrodes that are very close to the primary auditory cortex will be correlated to the audio waveform (Honey et al., 2012). Second, we will use an event-related potential (ERP) analysis to see an electrode’s response to the onset of a word.
[ ]:
# only run this cell in colab
!pip install nilearn mne mne_bids scipy pandas matplotlib
[1]:
import mne
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy import signal
from scipy.io import wavfile
from scipy.stats import pearsonr, zscore
from mne_bids import BIDSPath
from functools import partial
from nilearn.plotting import plot_markers
Just like the first tutorial, we will load the preprocessed neural activity in the high-gamma band for one subject using MNE.
[2]:
bids_root = "" # if using a local dataset, set this variable accordingly
file_path = BIDSPath(root=f"{bids_root}derivatives/ecogprep",
subject="03", task="podcast", datatype="ieeg", description="highgamma",
suffix="ieeg", extension=".fif")
print(f"File path within the dataset: {file_path}")
# You only need to run this if using Colab (i.e. if you did not set bids_root to a local directory)
if not len(bids_root):
!wget -nc https://s3.amazonaws.com/openneuro.org/ds005574/$file_path
file_path = file_path.basename
File path within the dataset: ../../monkey/derivatives/ecogprep/sub-03/ieeg/sub-03_task-podcast_desc-highgamma_ieeg.fif
However, this time after we read the file, we are going to select only a subset of electrodes. We do this simply to make the computations quicker and compatiable with fewer resources. The electrodes we chose to keep are known as “grid” electrodes because they’re arranged as a grid on the brain. In this case, we know which electrodes these are because of their names: they either start with LGA or LGB.
[3]:
raw = mne.io.read_raw_fif(file_path, verbose=False)
picks = mne.pick_channels_regexp(raw.ch_names, "LG[AB]*")
raw = raw.pick(picks)
raw
[3]:
| General | ||
|---|---|---|
| Filename(s) | sub-03_task-podcast_desc-highgamma_ieeg.fif | |
| MNE object type | Raw | |
| Measurement date | 2019-03-11 at 10:54:21 UTC | |
| Participant | sub-03 | |
| Experimenter | Unknown | |
| Acquisition | ||
| Duration | 00:29:60 (HH:MM:SS) | |
| Sampling frequency | 512.00 Hz | |
| Time points | 921,600 | |
| Channels | ||
| ECoG | ||
| Head & sensor digitization | 238 points | |
| Filters | ||
| Highpass | 70.00 Hz | |
| Lowpass | 200.00 Hz | |
And then, we will extract the underlying data to use directly:
[4]:
ecog_data = raw.get_data()
print(f"ECoG data has a shape of: {ecog_data.shape}")
ECoG data has a shape of: (235, 921600)
Correlation between electrode activity and audio stimulus¶
Next, we will load the raw audio waveform of the podcast. Note that the audio has a different sampling rate than the ECoG data, thus has many more time points.
[5]:
# Download the audio, if required
audio_path = f"{bids_root}stimuli/podcast.wav"
if not len(bids_root):
!wget -nc https://s3.amazonaws.com/openneuro.org/ds005574/$audio_path
audio_path = "podcast.wav"
[6]:
audio_sf, audio_wave = wavfile.read(audio_path)
print(f"Audio waveform is sampled at {audio_sf} Hz and has a shape of: {audio_wave.shape}")
# We will arbitrarily select the first channel from the stereo audio
if audio_wave.ndim > 1:
audio_wave = audio_wave[:, 0]
Audio waveform is sampled at 44100 Hz and has a shape of: (79380000, 2)
/tmp/ipykernel_3294141/641169259.py:1: WavFileWarning: Chunk (non-data) not understood, skipping it.
audio_sf, audio_wave = wavfile.read(audio_path)
You can also listen to the first 30 seconds of the podcast:
[7]:
from IPython.display import Audio
Audio(audio_wave[:audio_sf*30], rate=audio_sf)
[7]:
Following the procedure in Honey et al., 2012, we will extract the power modulations from the raw waveform between 200 and 5000 Hz. Then, we downsample this new time-series to the same sampling rate as the ECoG data (512 Hz). The entire process is encapsulated in the following function. It is not critical for you to follow it closely if you are not familiar with digital signal processing.
[8]:
def preprocess_raw_audio(x, fs, to_fs, lowcut=200, highcut=5000):
# See https://scipy-cookbook.readthedocs.io/items/ButterworthBandpass.html
def butter_bandpass(lowcut, highcut, fs, order=5):
nyq = 0.5 * fs
low = lowcut / nyq
high = highcut / nyq
b, a = signal.butter(order, [low, high], btype="band")
return b, a
def butter_bandpass_filter(data, lowcut, highcut, fs, order=5):
b, a = butter_bandpass(lowcut, highcut, fs, order=order)
y = signal.lfilter(b, a, data)
return y
assert x.ndim == 1
# Step 1. Bandpass the high quality audio
y = butter_bandpass_filter(x, lowcut, highcut, fs, order=5)
# Step 2. Downsample to same freq as clinical system
# Number of new samples is N = n * (to_fs / fs)
y = signal.resample(y, num=round(x.size / fs * to_fs))
# Step 3. Take audio envelope
envelope = np.abs(signal.hilbert(y - y.mean()))
return envelope
Notice that after preprocessing, the audio power has a new sampling rate and the same number of samples as the ECoG data:
[9]:
ecog_sr = raw.info['sfreq']
audio_power = preprocess_raw_audio(audio_wave, audio_sf, ecog_sr)
print(f"Audio power is sampled at {ecog_sr} Hz and has a shape of: {audio_power.shape}")
Audio power is sampled at 512.0 Hz and has a shape of: (921600,)
Now we have both time series that we want to correlate. We will loop through each electrode at a time, and correlate its neural activity with the audio power, saving the result in a list.
[10]:
n_electrodes = ecog_data.shape[0]
correlations = np.zeros(n_electrodes)
for i in range(len(ecog_data)):
result = pearsonr(audio_power, ecog_data[i])
correlations[i] = result.statistic
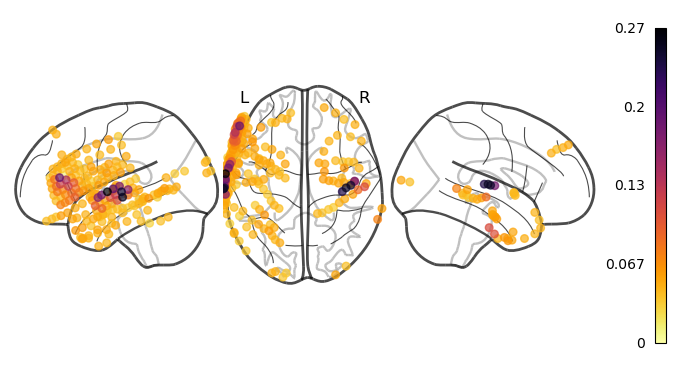
Great! We computed the correlations, now what? Since we have one correlation per electrode, we can view the results directly on the brain. This way, we verify that electrodes located near the primary auditory cortex do show a higher correlation to the audio stimulus than other electrodes.
[11]:
ch2loc = {ch['ch_name']: ch['loc'][:3] for ch in raw.info['chs']}
coords = np.vstack([ch2loc[ch] for ch in raw.info['ch_names']])
coords *= 1000 # nilearn likes to plot in meters, not mm
print("Coordinate matrix shape: ", coords.shape)
# order will ensure that the highest correlations are plotted on top (last)
order = correlations.argsort()
plot_markers(correlations[order], coords[order],
node_size=30, display_mode='lzr',
node_vmin=0, node_cmap='inferno_r', colorbar=True)
plt.show()
Coordinate matrix shape: (235, 3)
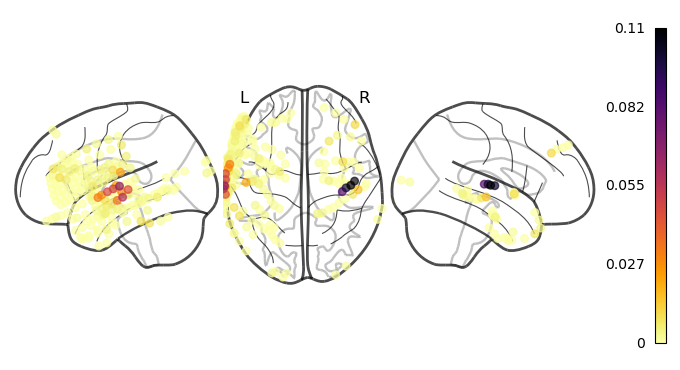
This example made a strong simplifying assumption: that the audio and electrode activity are synchronized simulatenously. However, this does not need be true for all electrodes. There are some electrodes who will be processing the incoming signal and thus will have some delay. To account for this possiblity, a cross correlation analysis is particularily usseful.